Introduction
Visit H3Africa.org for more details.
H3Africa Biospecimen and Data Catalogue
The H3Africa Catalogue offers researchers the ability to search information about H3Africa studies. As part of funder requirements, H3Africa samples are deposited in three H3Africa biorepositories and genomic data is submitted to public repositories such as the European Genome-phenome Archive (EGA) or the European Nucleotide Archive (ENA). The objective is to facilitate further research that will benefit study participants. Access to data and biospecimens is controlled by the H3Africa Data and Biospecimen Access Committee (DBAC). More information on the H3Africa policies for data and biospecimen access and release can be found here: H3Africa policy documents
Information in the H3Africa catalogue is derived from the H3Africa Archive hosted by H3ABioNet and the three biorepositories, which are based in Uganda, Nigeria and South Africa:
The H3Africa archive is a custodian of the H3Africa genomic and phenotype data and prepares the data for submission to the EGA or ENA. Metadata from the archive and biorepositories are submitted to the catalogue monthly to enable users to search:
- Study - by study name, design or disease
- Participant type - by sex, ethnicity or case/control
- Biospecimen - by country of origin, type or feature
From search results users can submit a request for access to the DBAC.
Overview of the catalogue architecture
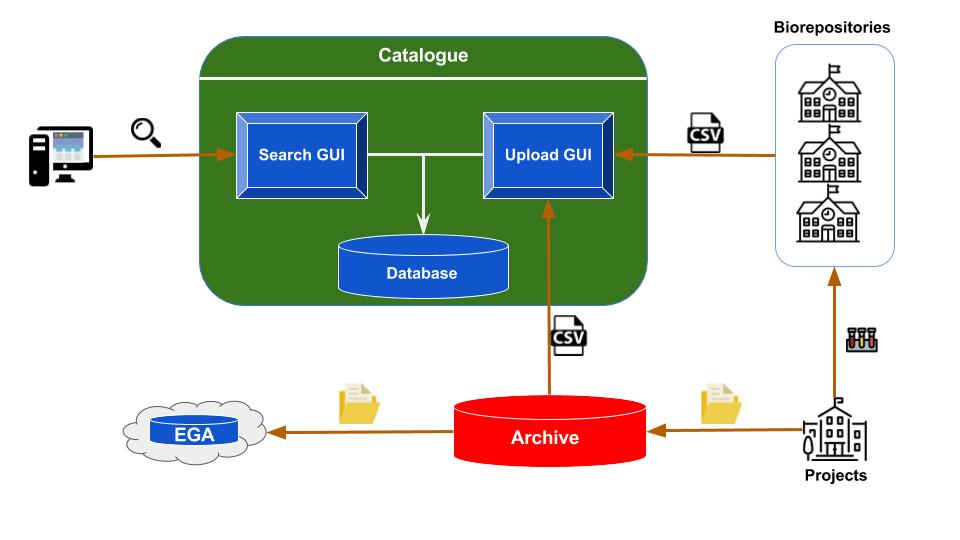 Figure 1: Overview of the catalogue architecture. At the bottom right are the institutions running the studies,
collecting the biospecimens, processing them and generating genomic and phenotype data. A portion of the biospecimens,
along with their metadata are sent to one of the biorepositories. The biorepositories update the content of their LIMS
with the new information. The genomic datasets generated from the studies are sent to the Archive, where they are held
for a period of 9 months before submission to public repositories. The Archive and biorepositories export csv files
with metadata for each project and upload them into the catalogue, triggering an update of the catalogue database.
Therefore, the catalogue end users are provided with the latest information in the Archive and biorepositories
Figure 1: Overview of the catalogue architecture. At the bottom right are the institutions running the studies,
collecting the biospecimens, processing them and generating genomic and phenotype data. A portion of the biospecimens,
along with their metadata are sent to one of the biorepositories. The biorepositories update the content of their LIMS
with the new information. The genomic datasets generated from the studies are sent to the Archive, where they are held
for a period of 9 months before submission to public repositories. The Archive and biorepositories export csv files
with metadata for each project and upload them into the catalogue, triggering an update of the catalogue database.
Therefore, the catalogue end users are provided with the latest information in the Archive and biorepositories
The catalogue is a Java application written in a Spring boot framework. Three modules compose the catalogue: an upload module, a database and a search engine. The upload module expects a well formatted csv file structured following predefined templates. There is one template for the repositories and two for the Archive, which splits the information into project and participant metadata. The template restricts the nomenclature of the captured field to ensure data harmonization. The export and uploading of csv files is not necessarily the best option for updating the catalogue database, an Application Programming Interface (API) would be more appropriate. However, none of the systems running in the biorepositories or the Archive have implemented such a solution yet.
The metadata in the catalogue are stored in a Neo4j graph, schemaless and NoSQL database. In a graph database, nodes represent the entities that may be extracted from the metadata, e.g Study, Participant, Biospecimen, etc. and the vertices represent the logic relating the nodes to each other, e.g a Biospecimen node is related to a Participant node.
The search engine module comes with a user friendly web interface with different HTML fields for filtering the content of the database. Although it is possible to query participant and biospecimen fields, the result is provided as a list of studies with general summary information about their participants and biospecimens. For ethical reasons, despite being anonymised, the participant and biospecimen IDs are hidden to protect the study individuals. Only authorized users, such as the DBAC, can view these when necessary.
Catalogue data capturing
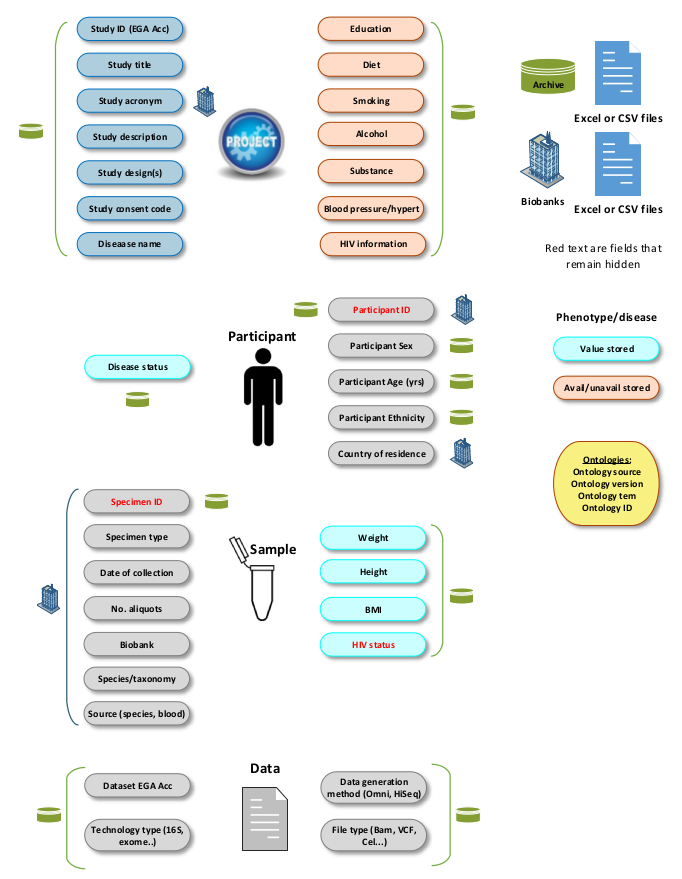 Figure 2:
All the CSV columns captured in the catalogue database. The columns are categorised into 4 principal entities: Project,
Participant, Biospecimen and Dataset. In addition, their provenance from Biorepositories or the Archive, is shown.
Sensitive information is shown in red meaning that this is hidden and not shown to the end users.
Figure 2:
All the CSV columns captured in the catalogue database. The columns are categorised into 4 principal entities: Project,
Participant, Biospecimen and Dataset. In addition, their provenance from Biorepositories or the Archive, is shown.
Sensitive information is shown in red meaning that this is hidden and not shown to the end users.
To ensure consistency in the information received from the biorepositories, a standard CSV template is used as a model for export of biospecimen metadata from the LIMS (Figure 2). Two standard CSV templates are defined for receiving data from the Archive.
Biospecimen data comes from the biorepositories, while Study, Participant and Dataset information originates from the Archive. However, some common fields exist between the biorepositories and Archive, and play a key role in linking the information, e.g the Study acronym and Participant ID links a biospecimen in a biobank to a study and dataset in the archive.